Projects
UV, NER and chromatin
The ultraviolet (UV) component of sunlight causes DNA damage that, if not repaired, can lead to mutations and increased risk for skin cancer. UV light induces two major types of damage: cyclobutane pyrimidine dimers (CPDs; 70-80 % of total damage) and (6-4) pyrimidine-pyrimidone dimers [(6-4)PD; 20-30 % of total damage]. CPDs are formed by covalent bonds between two carbon atoms (C-5 and C-6) of adjacent pyrimidines in the same DNA strand, forming a cyclobutyl ring; (6-4)PDs are formed by a covalent bond between the C-6 position of one pyrimidine and theC-4 position of the adjacent pyrimidine.
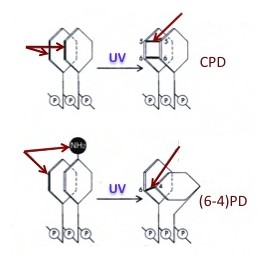
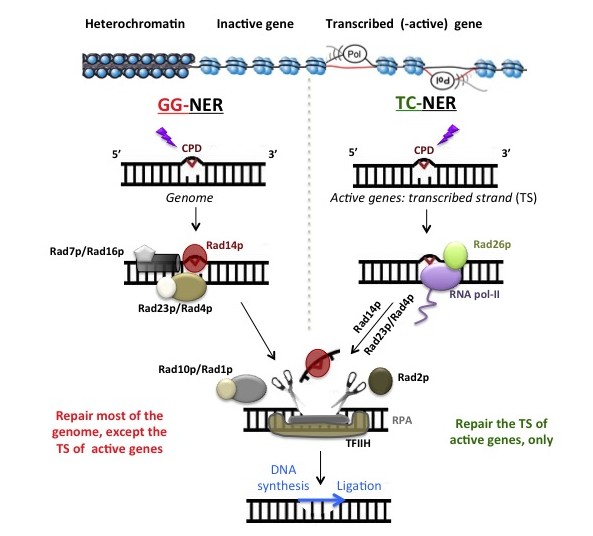
In eukaryotes, the genome is organized into nuclear domains that have distinct chromatin structures and functions: highly repetitive sequences, centromeres, telomeres, non-coding sequences, inactive genes, RNA polymerase-I, II and -III transcribed genes. Like DNA transcription and replication, NER is modulated by the structure of chromatin, and it is plausible that the kinetics of DNA repair vary among domains, suggesting that the mutation rate differs within nuclear domains.
rRNA genes chromatin
How modifications of histone proteins and chromatin remodeling affect NER in vivo is a challenging question (see UV, NER and chromatin). Therefore, most of the current knowledge has been gained by in vitro studies. We employ the yeast ribosomal genes (rRNA genes, or rDNA) as biological model to help answer some of the questions related to how NER operates on chromatin, in vivo.
The rRNA genes are highly expressed in growing cells, like cancer cells, and some agents used in chemotherapy target rRNA genes and their transcription rate. The rRNA genes are present in multiple copies that are housed in the nucleolus. They are of two distinct types: one that is permissive to transcription and the other that is transcription refractive. Synthesis of rRNA can be regulated by three different mechanisms: (1) designation of the total number of active genes, (2) modulation of the transcription initiation rate and, (3) regulation of RNA polymerase I (RNAPI) elongation rate.
Nucleosomes are not present in the coding regions of rRNA genes that are active but are found on the same coding DNA sequences when genes are inactive. Similar to the inactive rDNA copies, nucleosomes are present on most of the DNA sequences flanking rRNA genes.
Cisplatin (Cis-diaminedichloroplatinum)
Many chemotherapeutic drugs generate DNA damage that block DNA replication and, or, transcription. This results in cell cycle arrest and apoptosis. However, cancer cells can avoid cell death by a number of mechanisms, whereby DNA repair is one of the most important.
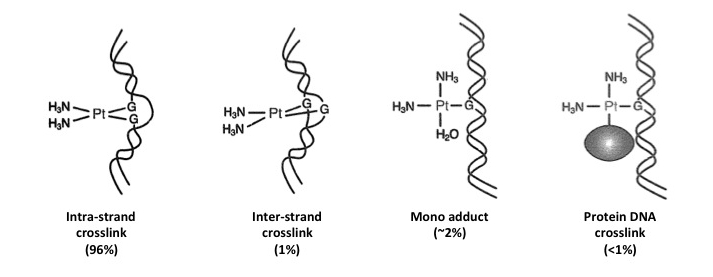
Cisplatin is used in chemotherapy to treat a number of tumors. Chlorine anions interact with hydroxy groups (mostly at N7 of purines), forming different types of DNA lesions: 1) intra-strand crosslinks (96%); 2) inter-strand crosslinks (1%); 3) mono adducts (~2%); 4) DNA-protein crosslinks (<1%).
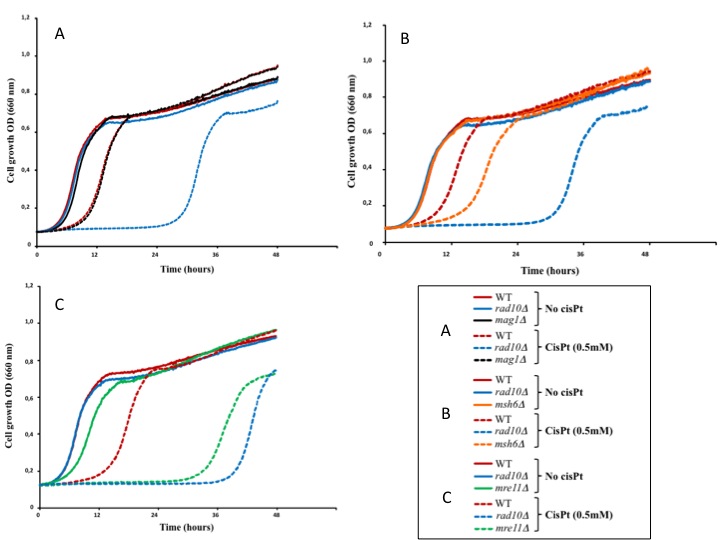
For example, the effect of cisplatin on cell growth was measured in different yeast strains: WT for DNA repair pathways, nucleotide excision repair (NER) deficient (rad10D), base excision repair (BER) deficient (mag1D), mismatch repair (MMR) deficient (msh6D) and homologous recombination repair (HR) deficient (mre11D) cells.
The cellular response to HU treatments has been considerably well analyzed in the yeast Saccharomyces cerevisiae. It was found that the inhibition of dNTP synthesis by HU triggered the S-phase checkpoint response. In turn, this promoted stabilization of stalled replication forks and blocked cell-cycle progression. Additionally, it was found that in HU treated yeast there were groups of genes that were induced and other that were repressed. Among the repressed genes there were those implicated in cell wall biogenesis, cytokinesis, bud site selection and cell polarity, or those implicated in regulation of mitosis, transcription and rRNA processing.
Transcription of rRNA genes and the production of mature rRNAs are required for cell growth and cell division. For example, in fast growing cancer cells rRNA genes are heavily transcribed.
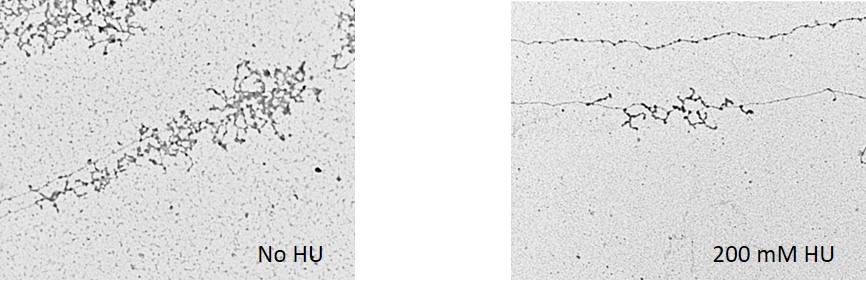
In yeast there are ~150 rRNA genes. From our studies on HU treated yeast, we concluded that most rRNA genes have an open form of chromatin, but only a portion of them is loaded with RNA polymerase I. Also, we found that the overall number of active promoters and RNA polymerase I-transcription activity remain largely unchanged after addition of HU to the media, indicating that in HU arrested cells rRNA genes stay ‘poised’ for transcription. However, despite of these characteristics favorable to transcription, rRNA synthesis is impaired. Hence, although the most important outcome of the HU response is the inhibition of DNA replication, it is conceivable that compromised rRNA synthesis could contribute to the slow-down of cell division, adding more significance to the use of HU in combined chemotherapy.
